Open source software for high-res microscopy
With their special microscopes, experimental physicists can already observe single molecules. However, unlike conventional light microscopes, the raw image data from some ultra-high resolution instruments first have to be processed for an image to appear. For the ultra-high resolution fluorescence microscopy that is also employed in biophysical research at Bielefeld University, members of the Biomolecular Photonics Group have developed a new open source software solution that can process such raw data quickly and efficiently.
The Bielefeld physicist Dr. Marcel Müller reports on this new open source software in the latest issue of Nature Communications published on 21th March.
Conventional light microscopy can attain only a defined lower resolution limit that is restricted by light diffraction to roughly 1/4 of a micrometre. High resolution fluorescence microscopy makes it possible to obtain images with a resolution markedly below these physical limits.
The physicists Stefan Hell, Eric Betzig, and William Moerner were awarded the Nobel Prize in 2014 for developing this important key technology for biomedical research. Currently, one of the ways in which researchers in this domain are trying to attain a better resolution is by using structured illumination.
At present, this is one of the most widespread procedures for representing and presenting dynamic processes in living cells. This method achieves a resolution of 100 nanometres with a high frame rate while simultaneously not damaging the specimens during measurement.
Such high resolution fluorescence microscopy is also being applied and further developed in the Biomolecular Photonics Group at Bielefeld's Faculty of Physics. For example, it is being used to study the function of the liver or the ways in which the HI virus spreads.
However, scientists cannot use the raw images gained with this method straight away. 'The data obtained with the microscopy method require a very laborious mathematical image reconstruction. Only then do the raw data recorded with the microscope result in a high-resolution image,' explains Professor Dr. Thomas Huser, head of the Biomolecular Photonics Group.
Because this stage requires a complicated mathematical procedure that has been accessible for only a few researchers up to now, there was previously no open source software solution that was easily available for all researchers. Huser sees this as a major obstacle to the use and further development of the technology. The software developed in Bielefeld is now filling this gap.
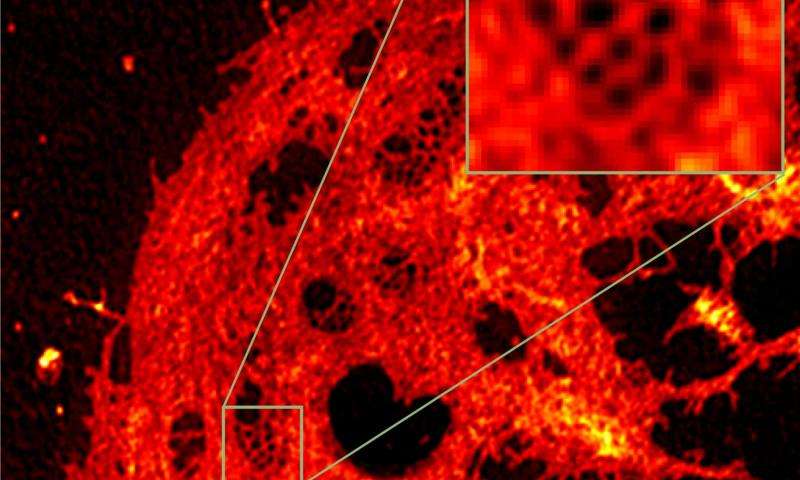
The images show a liver cell before and after processing the data with the software developed at Bielefeld University. Credit: Bielefeld University
Dr. Marcel Müller from the Biomolecular Photonics Group has managed to produce such universally implementable software. 'Researchers throughout the world are working on building new, faster, and more sensitive microscopes for structured illumination, particularly for the two-dimensional representation of living cells.
For the necessary post-processing, they no longer need to develop their own complicated solutions but can use our software directly, and, thanks to its open source availability, they can adjust it to fit their problems,' Müller explains.
The software is freely available to the global scientific community as an open source solution, and as soon as its availability was announced, numerous researchers, particularly in Europe and Asia, requested and installed it. 'We have already received a lot of positive feedback,' says Marcel Müller. 'That also reflects how necessary this new development has been.'





